Probiotics: Balancing the Dynamic Behaviour of Microbial Association and Intelligence
Dr. Hemant Purohit [Former Head, Environmental Biotechnology &/or Genomics Division, CSIR- National Environmental Engineering Research Institute (NEERI), Nagpur] is among the pioneering scientists who initiated the microbial genomics program in India. He is best known for applying microbial genomics tools in attaining basic knowledge to products in different models such as contaminated ecological niches, sewage/wastewater, plant rhizosphere, and the human gut.
His research interests lie in microbial genomics and designing Pre-, Pro- and Postbiotics for plants, animals, and humans. He is an alumnus of Nagpur University from 1986 where he finished his masters and doctorate degrees. This was followed by two postdoctoral projects at Dept of Biochemistry, University of Hull, UK and Lab Mol. Bio, NINDS, NIH, Bethesda, USA respectively. Subsequently, he joined as a Scientist in 1992 at CSIR-NEERI. He got promoted to Chief Scientist & Head for the Environmental Biotechnology &/or Genomics Division in 2003.
In his notable research career, he coordinated 35 projects which were funded by DBT, DST & CSIR & 21 projects were funded by industries. To cite a few:
- Application of microbial genomics tools: designing aerobic wastewater treatment and anaerobic food waste to biogas production; plant-microbe interactions and bioremediation (DBT/CSIR/Industry funded projects)
- Collecting and mining 2,00,000 bacteria for 4 bioactive therapeutic compounds (DBT funded multi-institutional project)
- Microbial diversity of human gut (ITC, Bengaluru)
- Clinical diagnostics and application of Ayurveda concepts in microbiome performance (in-house project)
- Public-private partnership DBT program with eight different institutions
He has to his credit over 300 publications with ~10,000 citations.
Microbes are vital participants in managing the energy flow for any ecological system. They are the primary receptors that recognize upcoming perturbances before they can be actually realized.
The association of microbes with the host system bring out the required biological performance of that ecosystem. The microbial community engages in a diverse role as per the demand of the ecosystem. For example, microbes control the nutrient flow in river sediment or polluted soil1 ; bridge the plants with a rhizospheric pool of biochemical intermediates2; or extend the physiological intelligence through various intermediates in animal/human gut3.
Different biological systems exhibit unique kinds of host- microbe interactions. They usually involve microbes providing supplementary metabolic support through the biochemical pathways to produce different intermediates or vital metabolites for the host. In turn, the host helps microbes with their nutritional requirements. These relationships possess a typical microbial community structure if the system is close to a steady state. However, with the dynamic nature of the host, the evolving scenarios force a shift in the diversity and overall functional complexity of microbial community.
For example,- In humans, it has been shown that the gut microbiome changes with age;
- The rhizosphere community at the time of seed germination is different than in matured plant;
- The selective enrichment of microbes occurs with changed biogeochemistry, and it correlates with seasonal variation in river sediments.
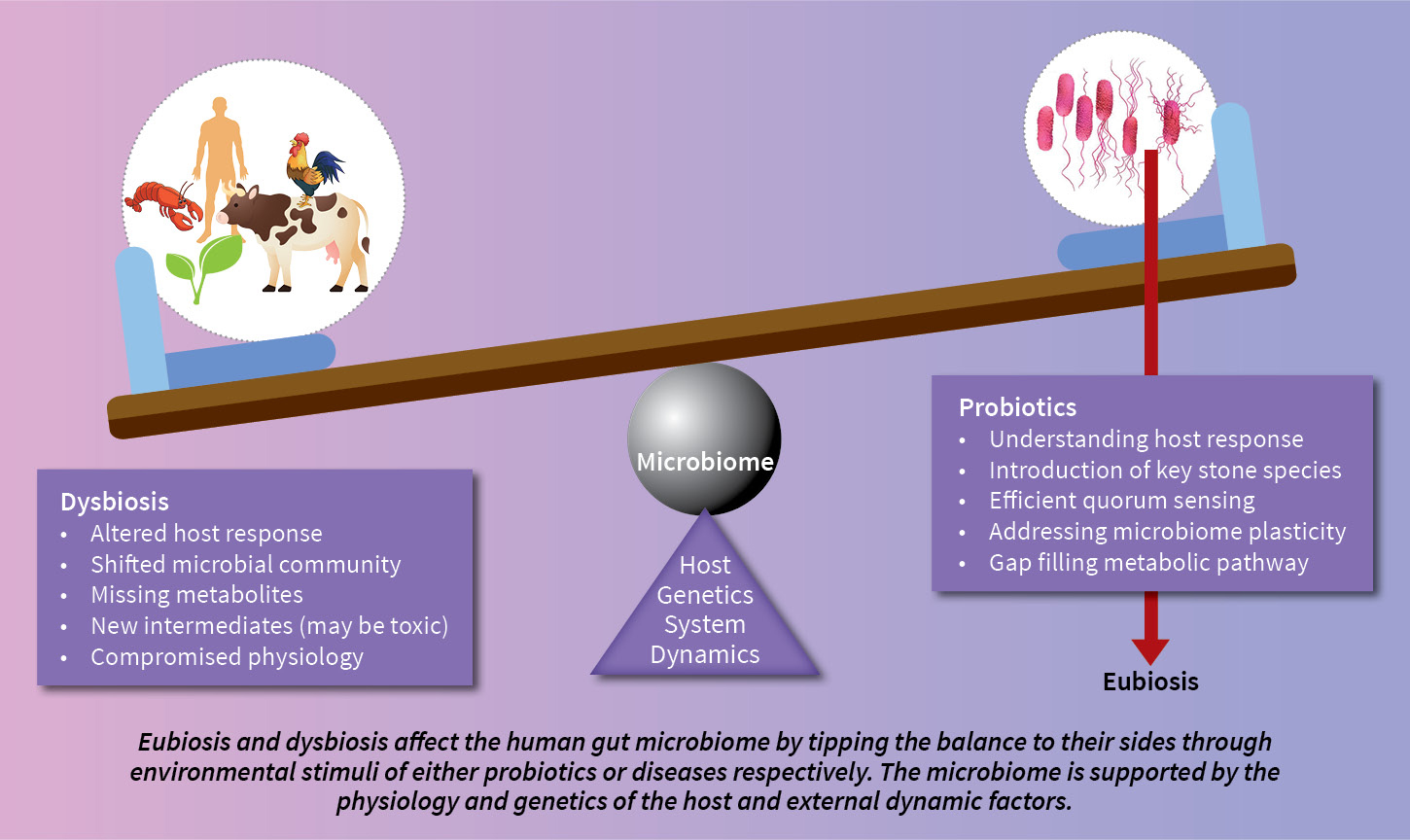
Let us understand these host-microbe associations with the human gut microbiome as an example. We can define this association as a fed-batch bioreactor, where with a typical lifestyle, for microbes, the host provides a set of raw materials and introduces various biochemicals. The residing microbial community in the gut, supports the host metabolism and generates a pool of intermediates depending on the raw material received. With age, the host changes its lifestyle and releases different sets of biochemicals as directed by its genetic constitution. These changes challenge the gut microbiome, and there starts a shift in microbial community structure. Any perturbances to healthy growth, survival, and association in the gut microbiome indicate dysbiosis (diarrhoea, irritable bowel syndrome, etc) which demand medical interventions. Such dysbiosis lead to affected metabolism in the host.
humans, numerous dysbioses such as IBD, Type 2 diabetes have been defined which are linked to several clinical disorders. To deal with these issues of altered biochemical capacities, probiotics are emerging as a solution. These probiotics as single bacterium species or bacterial consortia are selected based on observed dysbiosis; and can be supplemented to overpass the metabolic gap.
However, the observed shift in microbial community structure cannot be oversimplified since host genetics and lifestyle play major roles in affecting the overall status. Similar trends are also emerging with a “Compound Annual Growth Rate” of more than 5% in animal husbandry, poultry, and shrimp farming where probiotic supplement helps in stimulating the immune system and reproduction.
Managing the nutrients or pollutants in soil or aquatic system through bioaugmentation of microbes has been practiced for a long time for agricultural usage and bioremediation. Other than managing the nutrients through biofertilizers, now probiotic consortia are also being discovered to balance the rhizosphere community and supplementary flow of intermediates required for healthy plant growth. These probiotics are designed in such a way that they are plant- specific and improve the interaction of the plant with the soil microbial community.
The bioinformatics platforms like iLINCS provide a holistic view of sharing of physiological responsibilities in these interactions at every level from gene organizations to their expressions4. Tools like timeOmics are also now coming up that can decode and fetch the biochemical events and develop pathways by connecting the metabolic intermediates5. These strategies will help in selecting molecules or designing a formulation which can be used as prebiotic to support improved host microbiome interactions.
Probiotics are selected bacteria with required biochemical capacities has a tremendous future with wide applications from correcting the soil microbial community to an integral part of personalized medicine.
References:- Shade A. Microbiome rescue: directing resilience of environmental microbial communities. Current Opinion in Microbiology. 2023;72(102263):1-9. doi:10.1016/j. mib.2022.102263
- Ravelo-Ortega G, Raya-González J, López-Bucio J. Compounds from rhizosphere microbes that promote plant growth. Current Opinion in Plant Biology. 2023;73(102336). doi:10.1016/j. pbi.2023.102336
- Miyauchi E, Shimokawa C, Steimle A, Desai MS, Ohno H. The impact of the gut microbiome on extra-intestinal autoimmune diseases. Nature Reviews Immunology. 2022; 23:9-23. doi:10.1038/ s41577-022-007>27-y
- Pilarczyk M, Fazel-Najafabadi M, Kouril M, et al. Connecting omics signatures and revealing biological mechanisms with iLINCS. Nature Communications. 2022; 13(4678). doi:10.1038/ s41467-022-32205-3
- Bodein A, Scott-Boyer MP, Perin O, Lê Cao KA, Droit A. timeOmics: an R package for longitudinal multi-omics data integration. Wren J, ed. Bioinformatics. 2021; 38(2):577-579. doi:10.1093/ bioinformatics/btab664